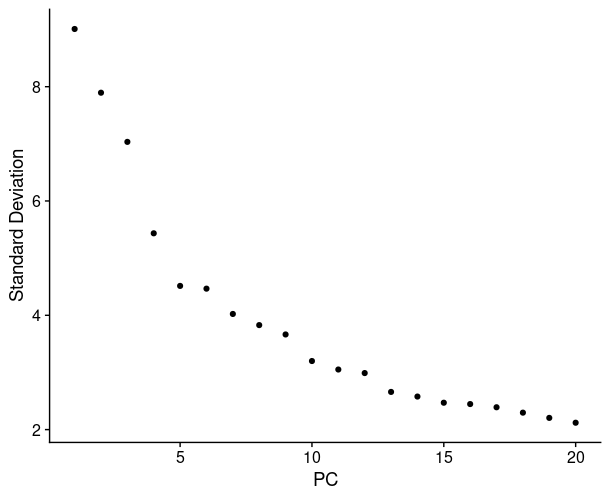
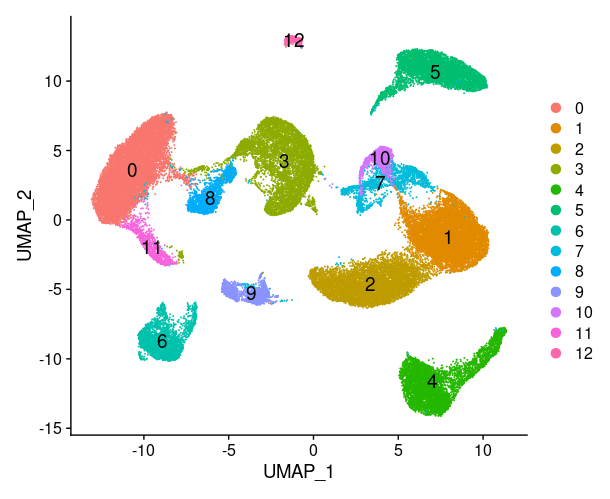
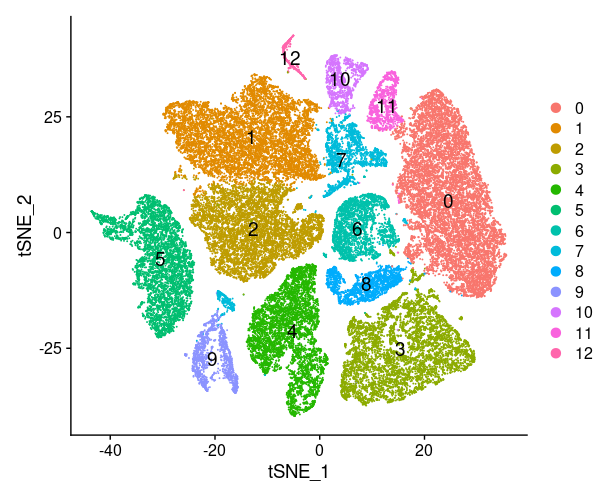
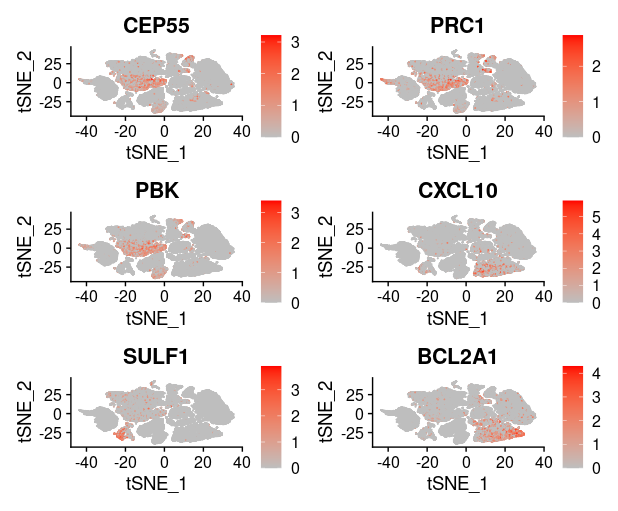
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
| siggene<-read.csv("TNBC_upregulation_gene.csv")
siggene<-siggene$x
siggene<-read.csv("TNBC_upregulation_gene.csv")
siggene<-siggene$x
plot_list = list()
for (i in 1:6) {
front=i*30-29
back=i*30
plot_list[[i]] = DotPlot(TNBC_scRNA, features = siggene[front:back]) + coord_flip()
print(plot_list[[i]])
}
KIAA0101<-c("L5", "NS5ATP9", "OEATC", "OEATC-1", "OEATC1", "PAF", "PAF15", "p15(PAF)",
"p15/PAF", "p15PAF")
EPT1<-c("SELI", "SEPI")
HN1<-c("ARM2", "HN1A")
DotPlot(TNBC_scRNA, features = HN1) + coord_flip()
select_siggene<-read.csv("first_filter.csv")
select_siggene<-select_siggene$candidate
plot_list = list()
for (i in 1:12) {
front=i*6-5
back=i*6
plot_list[[i]]=FeaturePlot(TNBC_scRNA, features = select_siggene[front:back],
reduction="tsne",
cols = c("grey", "red"))
print(plot_list[[i]])
}
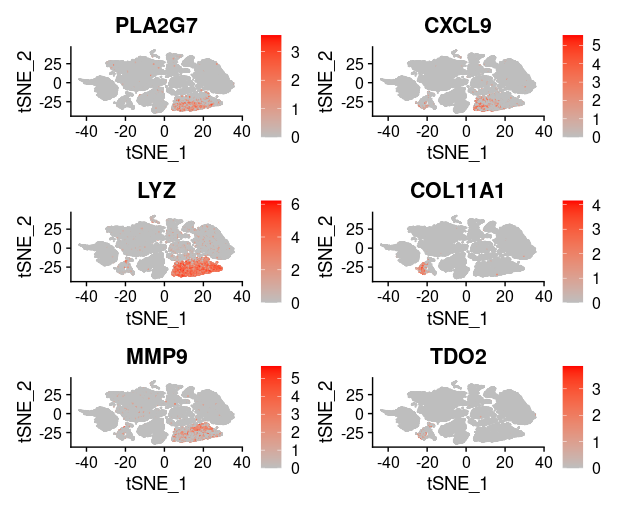
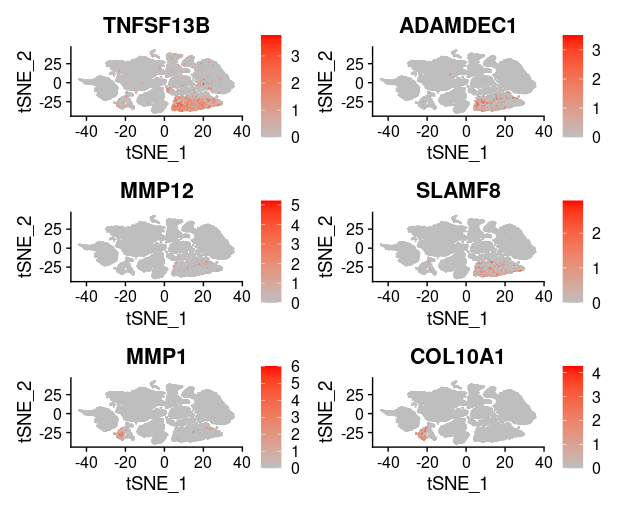
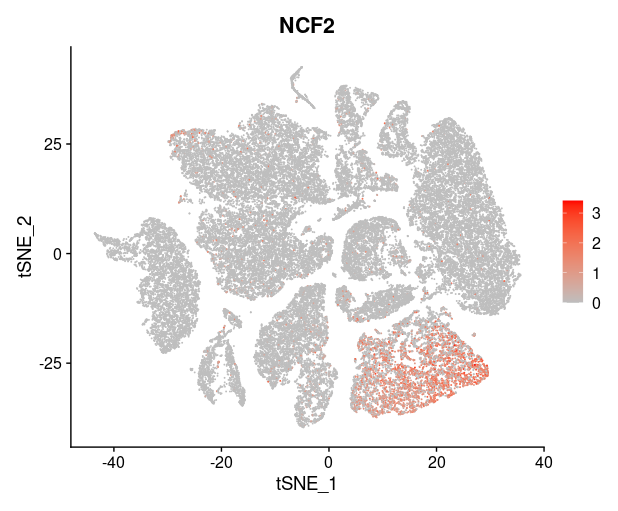
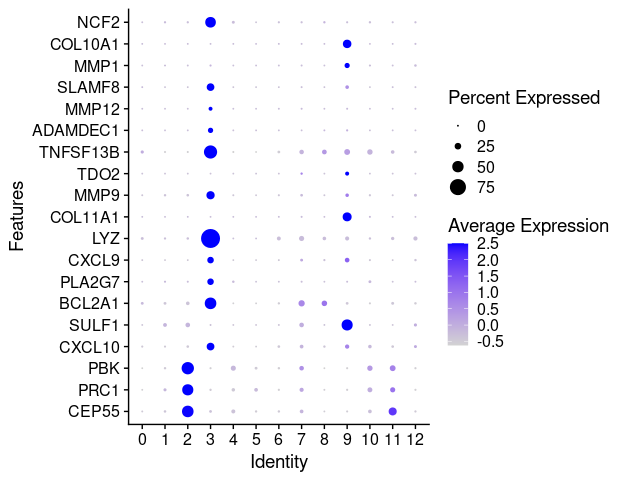
last_select_gene<-c("CEP55","PRC1","PBK","CXCL10","SULF1","BCL2A1",
"PLA2G7","CXCL9","LYZ","COL11A1","MMP9","TDO2",
"TNFSF13B","ADAMDEC1","MMP12","SLAMF8",
"MMP1","COL10A1","NCF2")
plot_list = list()
for (i in 1:4) {
front=i*6-5
back=i*6
plot_list[[i]]=FeaturePlot(TNBC_scRNA, features = last_select_gene[front:back],
reduction="tsne",
cols = c("grey", "red"))
print(plot_list[[i]])
}
p <- DotPlot(TNBC_scRNA, features = last_select_gene) + coord_flip()
p
|